scMultiNODE: Integrative and Scalable Framework for Multi-Modal Temporal Single-Cell Data
Abstract
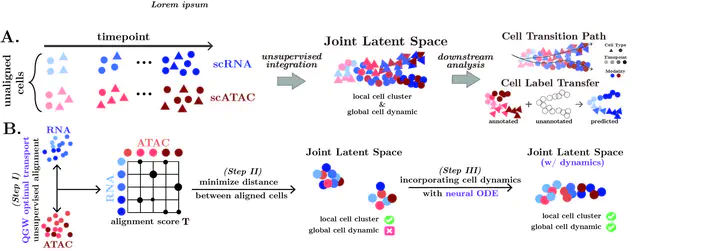
Measuring single-cell genomic profiles at different timepoints enables our understanding of cell development. This understanding is more comprehensive when we perform an integrative analysis of multiple measurements (or modalities) across various developmental stages. However, obtaining such measurements from the same set of single cells is resource-intensive, restricting our ability to study them jointly. We introduce scMultiNODE, an unsupervised integration model that combines gene expression and chromatin accessibility measurements in developing single cells, while preserving cell type variations and cellular dynamics. First, scMultiNODE uses a scalable, Quantized Gromov-Wasserstein optimal transport to align a large number of cells across different measurements. Next, it utilizes neural ordinary differential equations to explicitly model cell development with a regularization term to learn a dynamic latent space. Experiments on six real-world developmental single-cell datasets demonstrate that scMultiNODE can integrate temporally profiled multi-modal single-cell measurements more effectively than existing methods that focus on cell type variations and often overlook cellular dynamics. We also demonstrate that scMultiNODE’s joint latent space facilitates several insightful downstream analyses of single-cell development, including the investigation of complex cell trajectories and the enabling of cross-modal label transfer. The data and code are publicly available at https://github.com/rsinghlab/scMultiNODE.